- Home
- Genomes
- Genome Browser
- Tools
- Mirrors
- Downloads
- My Data
- Projects
- Help
- About Us
START AND STOP CODONS
Our genetic code is composed of DNA and, through the central dogma of biology, DNA is transcribed into RNA which is then translated into proteins. In the process of translation, not everything is translated, nor turned into usable genetic material. Many times, some RNA is spliced out before it's even translated. These sections of sequence are known as introns. Exons are the sections of RNA code that become translated into proteins. A way to remember this is, "exons are expressed" and introns are "in the trash".
Now, how does our RNA know what in our genetic code to include and what to exclude when creating proteins? This is where start and stop codons come into play. A codon is a sequence of three consecutive nucleotides that code for a specific amino acid. There are 20 different amino acids and 64 different codons. Each amino acid is encoded by either a single unique codon or multiple codons. For example, valine is encoded by four different codon sequences: GUU, GUA, GUC, and GUG. On the other hand, the amino acid methionine can only be coded for by AUG, the sole start codon (written as ATG in DNA).
To illustrate start and stop codons within our genetic code, we can use the Genome Browser to look for detailed examples (Figure 1). In this example, we will look at the gene METTL3, which can be found by typing in METTL3 into the search bar and clicking on the first drop-down result.
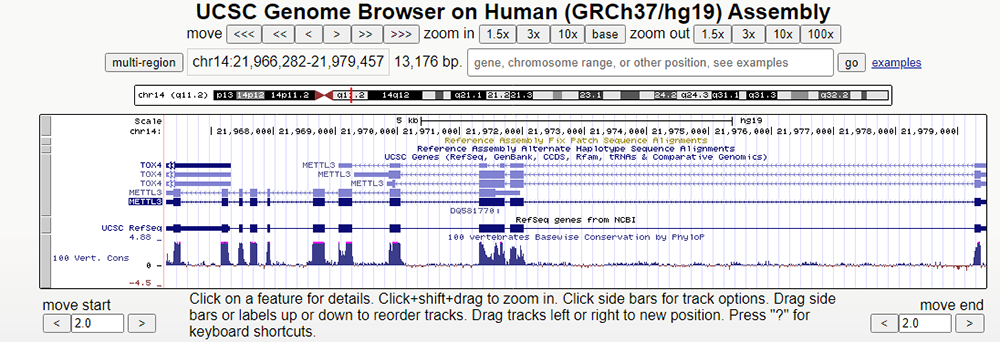
Figure 1. Main page of the METTL3 gene.
Here on the METTL3 main gene page, we can see a variety of data tracks already loaded in. Upon clicking on the gene's hyperlink (exon boxes), we learn, "This enzyme is involved in the posttranscriptional methylation of internal adenosine residues in eukaryotic mRNAs, forming N6-methyladenosine".
Now let's zoom in on the first exon of this gene (Figure 2). If we look at the direction of translation/arrows on the
introns, we can see that translation for this gene is going from right to left. Thus, the first exon will be the furthest to our right
(
 http://genome.ucsc.edu/s/education/mettl3_exon1):
http://genome.ucsc.edu/s/education/mettl3_exon1):
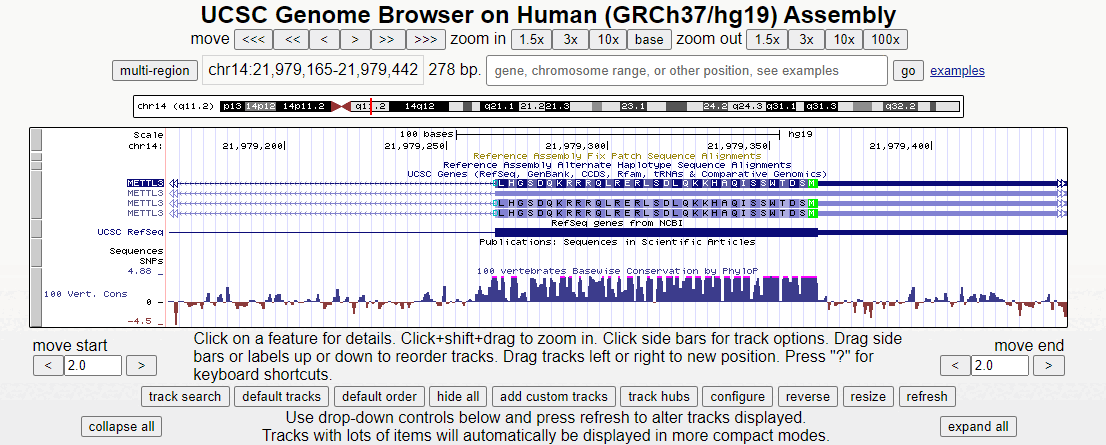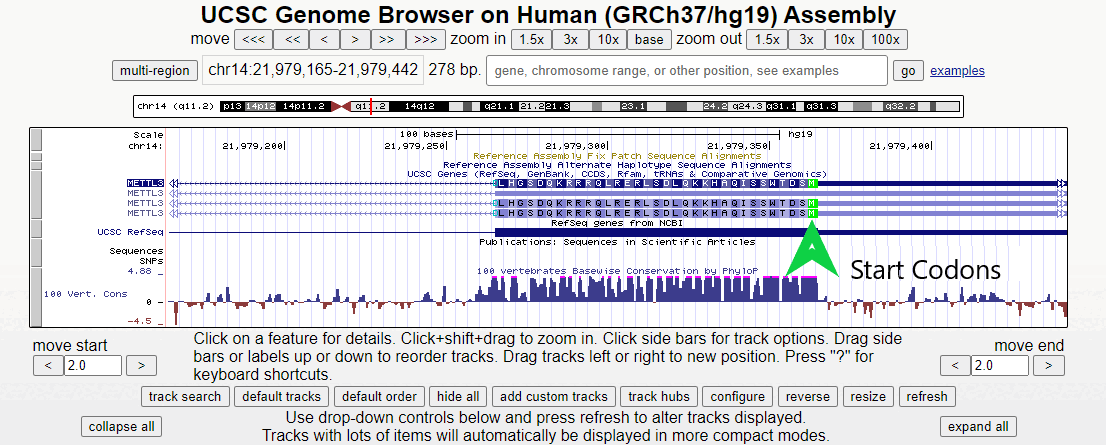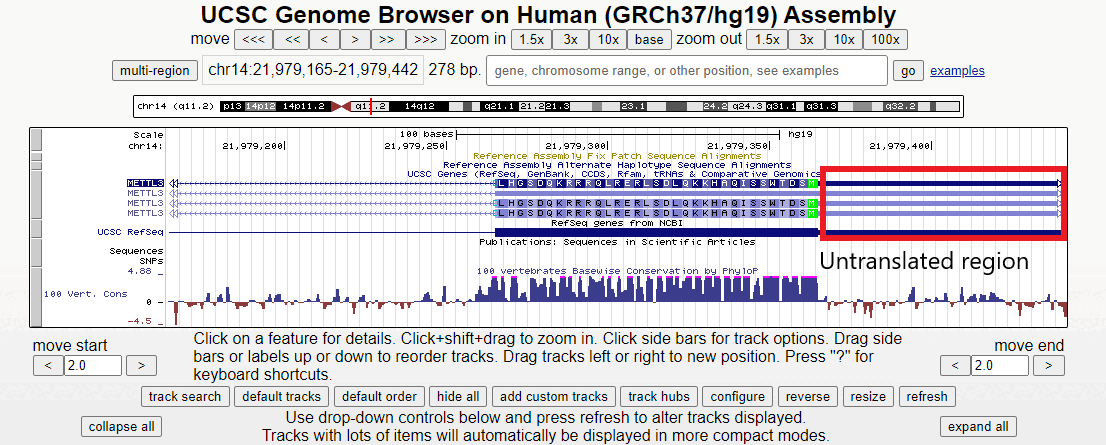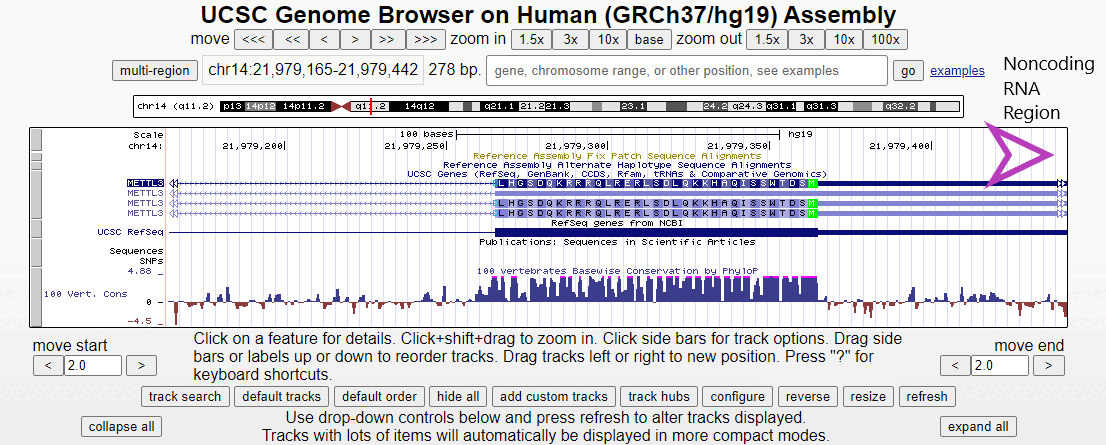
Figure 2. Short animation of the different regions surrounding the first exon of the METTL3 gene.
Zooming in here, the methionine start codon, colored in green, is observable. Notice how the sequence before the start codon is an untranslated region, depicted as a half-height box at the UTR region of the 5' end. Offscreen to the right is more non-coding RNA (indicated by double white arrows). Only after the start codon does translation actually begin in the right-to-left direction.
Now, let'ls scroll to the end of the METTL3 gene to look at the stop codons (Figure 3). There are three different stop codons: UAG, UAA, and UGA. These codons signal the end of the polypeptide chain during translation. These codons are also known as nonsense codons or termination codons as they do not code for an amino acid. Rather, they cause the release of the ribosome from the mRNA. Looking below, at the end of the last exon of the METTL3 gene, we can see the stop codon UAG
(
 http://genome.ucsc.edu/s/education/mettl3_stop):
http://genome.ucsc.edu/s/education/mettl3_stop):
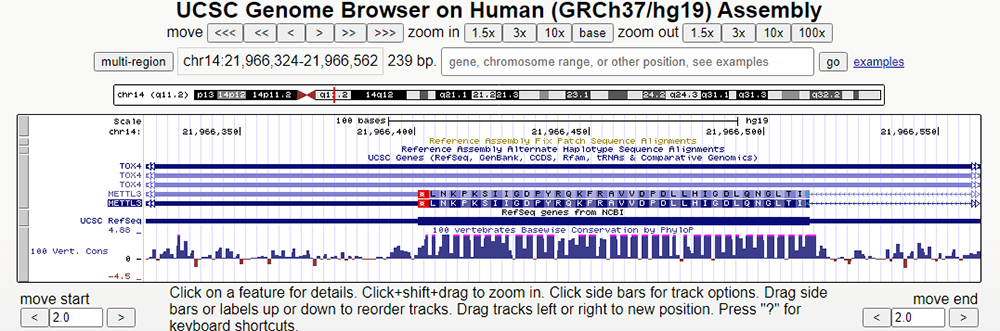
Figure 3. A stop codon (marked in red) found at the end of the METTL3 gene, initiating the end of translation. Below, we can observe how conservation decreases dramatically after the Stop codon in the "PhyloP" data track.
Notice in Figure 3 how after the stop codon, there are no further amino acids coded and translation ceases. In addition, if we look at the "100 vertebrates basewise conservation by PhyloP" data track a few tabs below, we can see that the conservation of nucleotides/codons drops off dramatically after the stop codon. However, where there are exons, or parts of the sequences that are expressed, the evolutionary conservation is high. This implies that conservation equates to functionality and that the exons are highly conserved as they are crucial to the functionality of the gene METTL3.
Written by Mateo Etcheveste, UCSC. Major: BS, Biomolecular Engineering