- Home
- Genomes
- Genome Browser
- Tools
- Mirrors
- Downloads
- My Data
- Projects
- Help
- About Us
MISSENSE SHORT VARIANTS
INTRODUCTION TO SHORT VARIANTS
Variants are found throughout all of our genomic sequences. In the past, the word "mutations" was typically used in place of "variants," but the present-day nomenclature is the latter, to avoid the negative assoications of the word. Some variants are unique to certain individuals or familial bloodlines ("private" variants), while others are common and well-documented. They can be completely innocuous, or result in faulty proteins with pathogenic ramifications. The Genome Browser can be utilized to visualize variants and their consequences.
A variant is typically defined as a deviation from the sequence of the reference assembly, which itself is the sequence of one arbitrary individual at any genomic location. The gene BRCA2 is known for having variants that are correlated with breast and ovarian cancer because its main function is as a tumor suppressor gene that helps repair damaged DNA; it also has a plethora of potential variations and serves as a good example for exploring different types of variants. We can examine some of these variations by turning on two types of "dbSNP" tracks, both of which tag variation sites in a gene. "SNP" is an acronym for "Single Nucleotide Polymorphism", and has been replaced by the more accurate "SNV" (Single Nucleotide Variant) to avoid misrepresenting the allele frequency — "polymorphism" implies that a variant is common in the population, which is not usually true. The database is still called dbSNP, however.
In Figures 1 and 2, the "UCSC Genes" track is displaying exon 11 of the BRCA2 gene. Below the "UCSC Genes" track are two "dbSNP" tracks, "dbSNP153" (upper) and "dbSNP151" (lower); these two tracks are different iterations of the same rsIDs and data, with some differences between the two data releases. The two tracks have different details available when clicking on one track or the other. Checking both versions of the rsID, or "reference SNP" can provide a wide range of insight into a variant.
Each colored box, labeled with an rsID, note marks a documented variation site within the sequence. A green box indicates that the variant is synonymous, or does not change the resulting amino acid. Red boxes signify that the nucleotide change does impact the amino acid sequence.

Figure 1. Zoomed-in picture of BRCA2, exon 11 (frame is 23 nucleotides across). The DNA sequence is at the top. The "UCSC Genes" track shows corresponding amino acids. The lower tracks labeled, "All Short Genetic Variants from dbSNP Release 153" and "Simple Nucleotide Polymorphisms (dbSNP 151)", have show identified variant sites.
An active Browser session for this view can be found at  https://genome.ucsc.edu/s/education/hg19_BRCA2variants
https://genome.ucsc.edu/s/education/hg19_BRCA2variants

Figure 2. How to enable the "dbSNP 153" and "dbSNP151" on human genome assembly hg19. The "UCSC Genes" track shows exon 11 of BRCA2.
MISSENSE SHORT VARIANTS
Figure 3 is an example of a missense variant, a single-nucleotide substitution that affects the resulting amino acid. The highlighted turquoise column intersects rs135936718 T/A/G, a location with three reported alleles (tri-allelic). While the reference genome allele is a T, this position has also been observed to also be an A or G, depending on the genome sequenced. We can use the strategy utilized to confirm the synonymous variant to corroborate that rs135936718 is in fact a missense variant, as its red rsID attests. The reference sequence is CAT, which codes for a histidine (or "H"). However, if the highlighted nucleotide in the reference codon is switched to one of its alternate alleles, an A or G, the sequence will read either CAA or CAG, which both correspond to the amino acid glutamine (or "Q"). This amino change confirms that rs135936718 is a potential missense variant site.
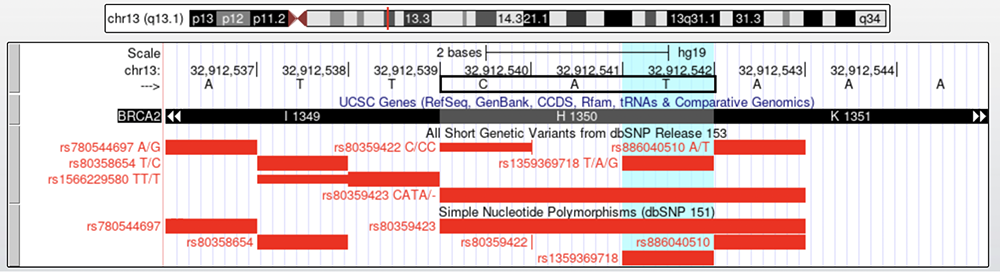
Figure 3.
(
 https://genome.ucsc.edu/s/education/hg19_BRCA2missense)
Highlighted in turquoise is rs135936718 T/A/G (T to A or G change), seen on both "dbSNP153" and "dbSNP 151."
Follow the highlight up to see the corresponding reference nucleotide (T). The black box on the DNA sequence marks the codon that includes the highlighted T allele.
Below, the "UCSC Genes Track" shows the amino acid (Histidine 1350) that is encoded by the codon boxed in black.
https://genome.ucsc.edu/s/education/hg19_BRCA2missense)
Highlighted in turquoise is rs135936718 T/A/G (T to A or G change), seen on both "dbSNP153" and "dbSNP 151."
Follow the highlight up to see the corresponding reference nucleotide (T). The black box on the DNA sequence marks the codon that includes the highlighted T allele.
Below, the "UCSC Genes Track" shows the amino acid (Histidine 1350) that is encoded by the codon boxed in black.
Clicking into rs135936718 will bring you to its details page, as seen in Figure 4. While some missense variations can have catastrophic and pathogenic effects, ClinVar, a database that collects data regarding medical consequences of genomic variations, marks this variant as having "uncertain-significance" (seen next to ClinVar). "Uncertain-significance" means that the alternate allele is not known to be either innocuous or pathogenic. Seeing as the alternate allele is "rare,i" stated as a note by UCSC, perhaps there is not enough data to support specific phenotypic consequences such as pathogenicity.

Figure 4. "dbSNP 153" details page of
 rs135936718.
Note ClinVar classification as uncertain-significance.
rs135936718.
Note ClinVar classification as uncertain-significance.
Figure 5 shows the "dbSNP 151" details page for rs135936718. Once again, "Coding annotations by dbSNP" and "UCSC's predicted function relative to selected gene tracks" provides confirmation of the same amino acid change deduced for ourselves in the previous paragraph.
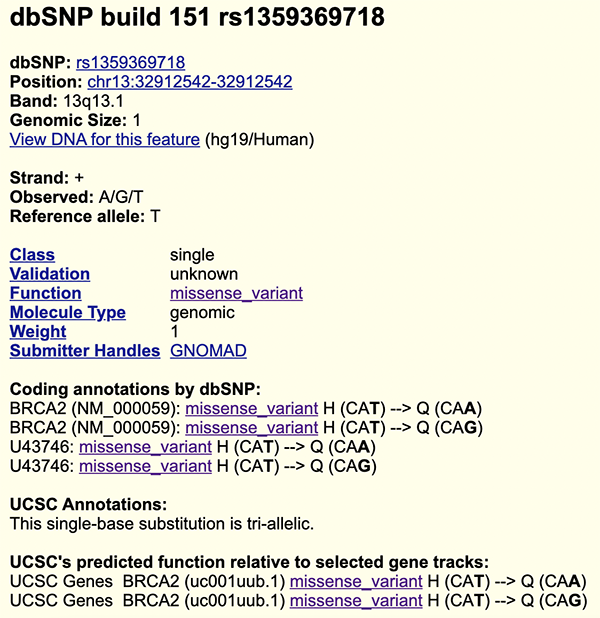
Figure 5. "dbSNP 151" details page of
 rs135936718. Note the histidine to glutamine (H > Q) amino acid changes for both T > A and T > G alleles.
rs135936718. Note the histidine to glutamine (H > Q) amino acid changes for both T > A and T > G alleles.
Written by Zoë Shmidt, UCSC. Major: BA, Biological Anthropology