- Home
- Genomes
- Genome Browser
- Tools
- Mirrors
- Downloads
- My Data
- Projects
- Help
- About Us
Data Information
Data last updated at UCSC: 2020-04-28
Track Description
Description
The NIH Genotype-Tissue Expression (GTEx) project was created to establish a sample and data resource for studies on the relationship between genetic variation and gene expression in multiple human tissues. This track shows median gene expression levels in 52 tissues and 2 cell lines, based on RNA-seq data from the GTEx final data release (V8, August 2019). This release is based on data from 17,382 tissue samples obtained from 948 adult post-mortem individuals.
Display Conventions
In Full and Pack display modes, expression for each gene is represented by a colored bargraph,
where the height of each bar represents the median expression level across all samples for a
tissue, and the bar color indicates the tissue.
Tissue colors were assigned to conform to the GTEx Consortium publication conventions.
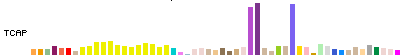
The bargraph display has the same width and tissue order for all genes.
Mouse hover over a bar will show the tissue and median expression level.
The Squish display mode draws a rectangle for each gene, colored to indicate the tissue
with highest expression level if it contributes more than 10% to the overall expression
(and colored black if no tissue predominates).
In Dense mode, the darkness of the grayscale rectangle displayed for the gene reflects the total
median expression level across all tissues.
The GTEx transcript model used to quantify expression level is displayed below the graph, colored to indicate the transcript class (coding, noncoding, pseudogene, problem), following GENCODE conventions.
Click-through on a graph displays a boxplot of expression level quartiles with outliers, per tissue, along with a link to the corresponding gene page on the GTEx Portal.
The track configuration page provides controls to limit the genes and tissues displayed, and to select raw or log transformed expression level display.Methods
Tissue samples were obtained using the GTEx standard operating procedures for informed consent and tissue collection, in conjunction with the National Cancer Institute Biorepositories and Biospecimen. All tissue specimens were reviewed by pathologists to characterize and verify organ source. Images from stained tissue samples can be viewed via the NCI histopathology viewer. The Qiagen PAXgene non-formalin tissue preservation product was used to stabilize tissue specimens without cross-linking biomolecules.RNA-seq was performed by the GTEx Laboratory, Data Analysis and Coordinating Center (LDACC) at the Broad Institute. The Illumina TruSeq protocol was used to create an unstranded polyA+ library sequenced on the Illumina HiSeq 2000 and HiSeq 2500 platforms to produce 76-bp paired end reads with a coverage goal of 50M (median achieved was ~82M total reads).
Sequence reads were aligned to the hg38/GRCh38 human genome using STAR v2.5.3a assisted by the GENCODE 26 transcriptome definition. The alignment pipeline is available here.Gene annotations were produced using a custom isoform collapsing procedure that excluded retained intron and read through transcripts, merged overlapping exon intervals and then excluded exon intervals overlapping between genes. Gene expression levels in TPM were called via the RNA-SeQC tool (v1.1.9), after filtering for unique mapping, proper pairing, and exon overlap. For further method details, see the GTEx Portal Documentation page.
UCSC obtained the gene-level expression files, gene annotations and sample metadata from the GTEx Portal Download page. Median expression level in TPM was computed per gene/per tissue.
Subject and Sample Characteristics
The scientific goal of the GTEx project required that the donors and their biospecimen present with no evidence of disease. The tissue types collected were chosen based on their clinical significance, logistical feasibility and their relevance to the scientific goal of the project and the research community. Summary plots of GTEx sample characteristics are available at the GTEx Portal Tissue Summary page.
Data Access
The raw data for the GTEx Gene expression track can be accessed interactively through the Table Browser or Data Integrator. Metadata can be found in the connected tables below.
- gtexGeneModelV8 describes the gene names and coordinates in genePred format.
- hgFixed.gtexTissueV8 lists each of the 53 tissues in alphabetical order, corresponding to the comma separated expression values in gtexGeneV8.
- hgFixed.gtexSampleDataV8 has TPM expression scores for each individual gene-sample data point, connected to gtexSampleV8.
- hgFixed.gtexSampleV8 contains metadata about sample time, collection site, and tissue, connected to the donor field in the gtexDonorV8 table.
- hgFixed.gtexDonorV8 has anonymized information on the tissue donor.
For automated analysis and downloads, the track data files can be downloaded from
our downloads server
or the JSON API.
Individual regions or the whole genome annotation can be accessed as text using our utility
bigBedToBed. Instructions for downloading the utility can be found
here.
That utility can also be used to obtain features within a given range, e.g.
bigBedToBed http://hgdownload.soe.ucsc.edu/gbdb/hg38/gtex/gtexGeneV8.bb -chrom=chr21
-start=0 -end=100000000 stdout
Data can also be obtained directly from GTEx at the following link: https://gtexportal.org/home/datasets
Credits
Statistical analysis and data interpretation was performed by The GTEx Consortium Analysis Working Group. Data was provided by the GTEx LDACC at The Broad Institute of MIT and Harvard.
References
GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020 Sep 11;369(6509):1318-1330. PMID: 32913098; PMC: PMC7737656
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013 Jun;45(6):580-5. PMID: 23715323; PMC: PMC4010069
Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, Compton CC, DeLuca DS, Peter-Demchok J, Gelfand ET et al. A Novel Approach to High-Quality Postmortem Tissue Procurement: The GTEx Project. Biopreserv Biobank. 2015 Oct;13(5):311-9. PMID: 26484571; PMC: PMC4675181
Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015 May 8;348(6235):660-5. PMID: 25954002; PMC: PMC4547472DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, Reich M, Winckler W, Getz G. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics. 2012 Jun 1;28(11):1530-2. PMID: 22539670; PMC: PMC3356847