Description
This track displays the ENCODE Registry of candidate cis-Regulatory Elements (cCREs)
in the human genome, a total of 926,535 elements identified and classified by the ENCODE Data
Analysis Center according to biochemical signatures.
cCREs are the subset of representative DNase hypersensitive sites across ENCODE and
Roadmap Epigenomics samples that are supported
by either histone modifications (H3K4me3 and H3K27ac) or CTCF-binding data.
The Registry of cCREs is one of the core components of the integrative level of the
ENCODE Encyclopedia of DNA Elements.
Additional exploration of the cCRE's and underlying raw ENCODE data is provided by the
SCREEN
(Search Candidate cis-Regulatory Elements) web tool,
designed specifically for the Registry, accessible by linkouts from the track details page.
The cCREs identified in the mouse genome are available in a companion track,
here.
Display Conventions and Configuration
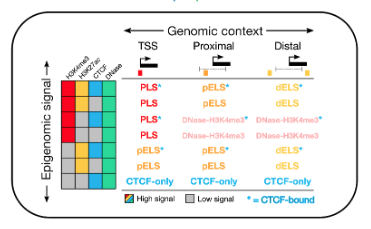
CCREs are colored and labeled according to classification by regulatory signature:
| Color |
|
UCSC label |
ENCODE classification |
ENCODE label |
| red |
prom |
promoter-like signature |
PLS |
| orange |
enhP |
proximal enhancer-like signature |
pELS |
| yellow |
enhD |
distal enhancer-like signature |
dELS |
| pink |
K4m3 |
DNase-H3K4me3 |
DNase-H3K4me3 |
| blue |
CTCF |
CTCF-only |
CTCF-only |
The DNase-H3K4me3 elements are those with promoter-like biochemical signature that
are not within 200bp of an annotated TSS.
Methods
All individual DNase hypsersensitive sites (DHSs) identified from 706 DNase-seq experiments
in humans (a total of 93 million sites from 706 experiments) were iteratively clustered
and filtered for the highest signal across all experiments, producing
representative DHSs (rDHSs), with a total of 2.2 million such sites in human.
The highest signal elements from this set that were also supported by high H3K4me3, H3K27ac
and/or CTCF ChIP-seq signals were designated cCRE's (a total of 926,535 in human).
Classification of cCRE's was performed based on the following criteria:
- cCREs with promoter-like signatures (cCRE-PLS) fall within 200 bp of an annotated GENCODE TSS
and have high DNase and H3K4me3 signals.
- cCREs with enhancer-like signatures (cCRE-ELS) have high DNase and H3K27ac with low H3K4me3
max-Z score if they are within 200 bp of an annotated TSS. The subset of cCREs-ELS within 2 kb
of a TSS is denoted proximal (cCRE-pELS), while the remaining subset is denoted distal
(cCRE-dELS).
- DNase-H3K4me3 cCREs have high H3K4me3 max-Z scores but low H3K27ac max-Z scores and do not
fall within 200 bp of a TSS.
- CTCF-only cCREs have high DNase and CTCF and low H3K4me3 and H3K27ac.
The GENCODE V24 (Ensembl 33) basic gene annotation set was used in this analysis.
For further detail about the identification and classification of ENCODE cCREs see
the About page of the
SCREEN web tool.
Data Access
The ENCODE accession numbers of the constituent datasets at the
ENCODE Portal
are available from the cCRE details page.
The data in this track can be interactively explored with the
Table Browser or the
Data Integrator.
The data can be accessed from scripts through our
API, the track name is "encodeCcreCombined".
For automated download and analysis, this annotation is stored in a bigBed file that
can be downloaded from
our download server.
The file for this track is called encodeCcreCombined.bb.
Individual regions or the whole genome annotation can be obtained using our tool
bigBedToBed which can be compiled from the source code or downloaded as a precompiled
binary for your system.
Instructions for downloading source code and binaries can be found
here.
The tool can also be used to obtain only features within a given range, e.g.
bigBedToBed http://hgdownload.soe.ucsc.edu/gbdb/hg38/encode3/ccre/encodeCcreCombined.bb -chrom=chr21 -start=0 -end=100000000 stdout
Release Notes
This annotation is based on ENCODE data released on or before September 14, 2018.
Data from the Common fund supported
Roadmap Epigenomics Mapping Consortium
(REMC) were included for building the ENCODE cCREs. Please see the 2015 paper on their analysis
of reference human genomes for more information.
Credits
This dataset was produced by the
ENCODE Data Analysis Center
(ZLab at UMass Medical Center). Please check the
ZLab ENCODE Public Hubs
for the most updated data.
Thanks to Henry Pratt, Jill Moore, Michael Purcaro, and Zhiping Weng, PI for providing
this data.
Thanks also to the ENCODE Consortium, the ENCODE production laboratories,
and the ENCODE Data Coordination Center for generating and processing the datasets used here.
References
ENCODE Project Consortium.
Expanded Encyclopedias of DNA Elements in the Human and Mouse Genomes.
Nature. 2020 July 30;583(7818):699-710
ENCODE Project Consortium.
An integrated encyclopedia of DNA elements in the human genome.
Nature. 2012 Sep 6;489(7414):57-74.
PMID: 22955616; PMC: PMC3439153
ENCODE Project Consortium.
A user's guide to the encyclopedia of DNA elements (ENCODE).
PLoS Biol. 2011 Apr;9(4):e1001046.
PMID: 21526222; PMC: PMC3079585
|