- Home
- Genomes
- Genome Browser
- Tools
- Mirrors
- Downloads
- My Data
- Projects
- Help
- About Us
ALCOHOL INTOLERANCE IN EAST ASIAN POPULATIONS
Ethanol, also known as drinking alcohol, is found throughout natural environments. Dating back to our very early primate days, humans would have been exposed to natural sources of ethanol such as fermenting fruits. Built within our bodies are processes that maintain our homeostatic state and are able to break down the alcoholic molecules that threaten to disrupt that state. In the course of digesting alcohol, our bodies make acetaldehyde, a molecule that can be poisonous at high levels and result in organ damage, impaired brain function, and other severe symptoms. Acetaldehyde is processed by an enzyme called acetaldehyde dehydrogenase, producing acetate; this enzyme is assembled when the gene ALDH2 is expressed. Therefore, in order to efficiently digest alcohol and the acetaldehyde that is produced in the process, one must possess a functioning ALDH2 gene. Figure 1 shows two isoforms of ALDH2 seen on the Genome Browser. Figure 2 shows the details page accessed by clicking into the gene, providing more information on the specific biomolecular processes ALDH2 and its resulting protein are involved in, along with some transcript numerical data such as exon count.
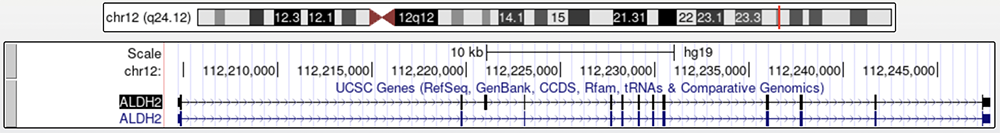
Figure 1. The ALDH2 gene region with "lUCSC Genes" track enabled.
Two isoforms are shown. Highlighted in black is the canonical transcript.
 https://genome.ucsc.edu/s/education/hg19_ALDH2
https://genome.ucsc.edu/s/education/hg19_ALDH2
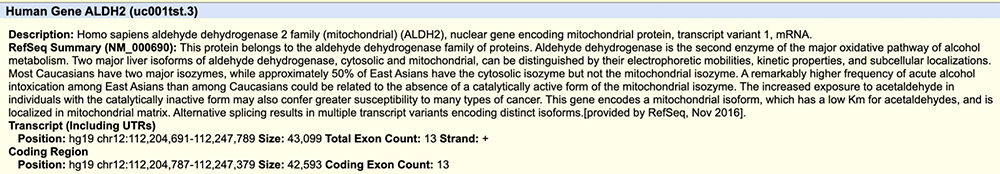
Figure 2. Part of the ALDH2 details page. Accessed by clicking on the gene.
 link.
link.
Because a functioning protein can only be produced by an intact gene, missense variants within ALDH2 can result in faulty protein production and decreased efficiency in digesting the acetaldehyde. These missense variants code for incorrect amino acids and disrupt the resulting polypeptide chain. The data track labeled "Simple Nucleotide Polymorphisms (dbSNP150)" can be turned on to see where common variants in ALDH2 lie (Figure 3); Note that this track shows all SNPs in the region, not just missense variants.
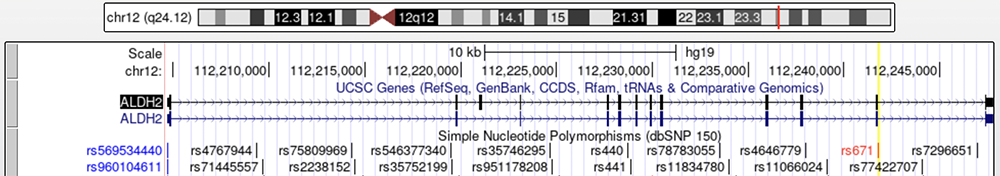
Figure 3. The "Simple Nucleotide Polymorphisms (dbSNP150)" track shows a variety
of variants that have been reported in ALDH2. The highlighted yellow column marks rs671, the variant associated with alcohol
flush syndrome.
 https://genome.ucsc.edu/s/education/hg19_ALDH2dbSNP150
https://genome.ucsc.edu/s/education/hg19_ALDH2dbSNP150
One missense variant, labeled rs671 (Figure 4), is the variant that causes "alcohol flush syndrome," a condition seen in approximately 30% to 50% of East Asian individuals (A smaller rsID number corresponds to an earlier entry, meaning that rs671 is one of the earliest uploaded into the database). This single nucleotide change creates an acetaldehyde dehydrogenase deficiency, meaning that alcohol consumption can result in a buildup of acetaldehyde that the body cannot process fast enough, resulting in acetaldehyde poisoning. The principal symptom of acetaldehyde poisoning is flushing, or erythema, on the face and body.
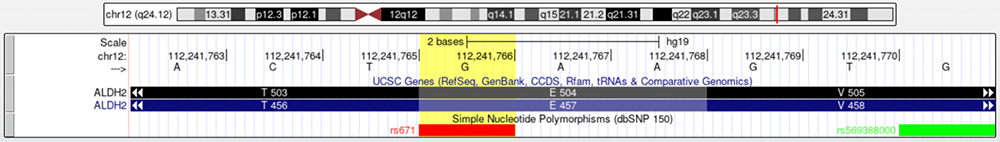
Figure 4. A zoomed in perspective of rs671, highlighted in Figure 3. The nucleotide base (G) and corresponding amino acids can be seen (E 504 and E457 in the two isoforms).
As usual, clicking into the details page of rs671 yields more information about the variant (Figure 5). This page provides information about the ancestral and derived allele; In this case, the "Observed" line shows that the nucleotide in this position has been documented as occurring as either an A or G. One can also see what alleles are observed in other primates ("Chimp allele", "Orangutan allele", "Macaque allele") in order to infer one variant as ancestral and the other as derived within the human lineage of simians. In this case, G would be considered the ancestral or "wild type", while A is the alternative or derived allele. Why this derived allele, with its pathogenic consequences, was positively selected for is still unknown. Not all derived alleles have pathogenic consequences, most do not, but this variant does. Going back to the main assembly of the gene, we can see that the reference sequence has a G in this position (Figure 4). This means that an individual who possesses the reference sequence as their own genome would not experience the symptoms of alcohol flush syndrome and could effectively break down acetaldehyde; An A allele would result in alcohol flush syndrome. If the individual is homozygous and has two copies of the A allele, the consequences of drinking alcohol would be severe; On the other hand, a heterozygous individual may be able to consume more alcohol while experiencing lesser symptoms (Wikipedia).

Figure 5. A section of the dbSNP150 details page for rs671.
 link.
link.
While the dbSNP track illuminated which allele is the derived (A) and which is the ancestral (G), imparting an assumption that the A allele results in alcohol flush syndrome, there are other tracks that can corroborate this and provide more information about the phenotypic consequences of the derived allele. GWAS, or the "Catalog of Published Genome-wide Association Studies", is a track that displays publications discussing phenotypes associated with a variant/rsID. Turning the GWAS track on while viewing ALDH2 shows that they also have data regarding rs671 (Figure 6).
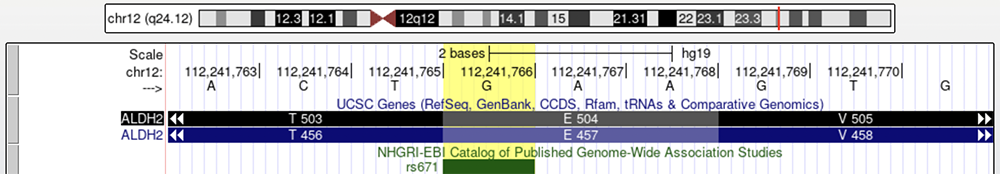
Figure 6. The GWAS track is turned on and the variant in question is highlighted in yellow.
 https://genome.ucsc.edu/s/education/hg19_ALDH2gwas
https://genome.ucsc.edu/s/education/hg19_ALDH2gwas
Clicking into rs671's entry in the GWAS track brings up a list of publications; There are 40+ publications relating to this variant. A majority of the publications are studies of cross-populational data that emphasize a high frequency of the derived allele found within East Asian populations. Figure 7 showcases one of these publications.

Figure 7. Information from one publication on the GWAS details page for rs671.
 link.
link.
The OMIM, or "Online Mendelian Inheritance in Man", data track can also be enabled in order to see their data on this variant (Figure 8). OMIM assigns their own identifiers to various alleles, with the first six digits referring to the gene; OMIM refers to rs671 as 100650.0001.
Figure 8. The OMIM track is turned on and the variant in question is highlighted in yellow.
 https://genome.ucsc.edu/s/education/hg19_ALDH2omim
https://genome.ucsc.edu/s/education/hg19_ALDH2omim
Clicking into the variant'ls link in the OMIM track will lead you to another details page (Figure 9). This page labels the variant as being associated with "acute alcohol sensitivity." It also provides the "Amino Acid Replacement" consequence of having the derived allele, A, rather than the ancestral or wildtype allele, G; Instead of possessing the codon GAA which corresponds to the amino acid glutamate, an individual with this mutation will have the codon AAA, which codes for a lysine amino acid (GLU540LYS). This phenomenon is referred to as a missense variant, or an allele change that encodes an altered polypeptide chain and results in a flawed protein; This explains why having the derived allele, A, results in acetaldehyde dehydrogenase enzymes that do not properly process acetaldehyde.

Figure 9. The Genome Browser's OMIM details page on rs671, labeled as 100650.0001 here.
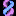 Link.
Link.
Clicking on "100650.0001" will take you to a more in-depth details page found on OMIM's website (Figure 10) which describes the variant as associated with esophageal cancer, alcohol-related.

Figure 10. OMIM's detail page for variant rs671/100650.0001 on their own website: https://omim.org/entry/100650#0001. Clicking the link "rs671" at the bottom allows you navigate back to UCSC (albeit to the hg38 assembly).
Turning on the ClinVar track and viewing ALDH2 on the Genome Browser further confirms this variant's pathogenicity (Figure 11). Take note of the red bead (which indicates a pathogenic variant) and the phenotypic consequences mentioned on the ClinVar details page, accessed if you click into the variant's gray box (Figure 12). The details page lists that some of the pathogenic phenotypes rs671 has been linked to are alcohol sensitivity, esophageal cancer, and others.
Figure 11. The ClinVar track is enabled and as information regarding rs671. Pathogenicity is implied via the red bead.
 https://genome.ucsc.edu/s/education/hg19_ALDH2clinvar
https://genome.ucsc.edu/s/education/hg19_ALDH2clinvar
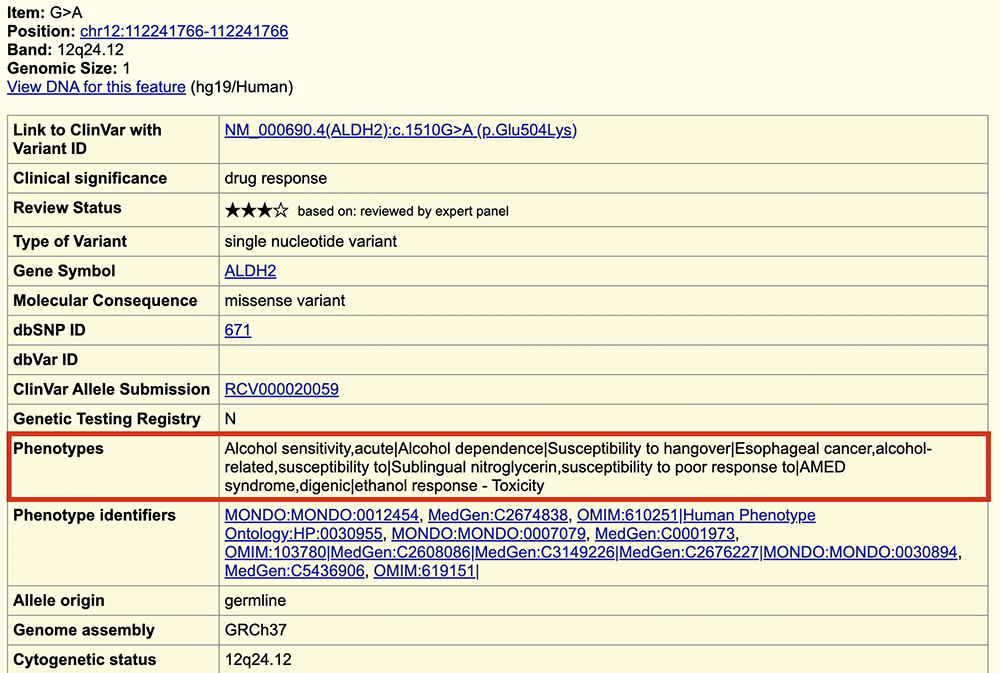
Figure 12. The ClinVar details page for rs671. Note the associated phenotypes, boxed in red.
 link.
link.
Earlier, many of the publications within the GWAS details page mentioned that this variant is associated with certain populations (Figure 7). We will enable the gnomAD ("Genome Aggregation Database") and HGDP ("Human Genome Diversity Project") tracks, two datasets that provide information about a variant'ls population frequencies, and see if we can use the Genome Browser to confirm rs671's high frequency in East Asian populations. Figure 13 shows that gnomAD has data regarding rs671.
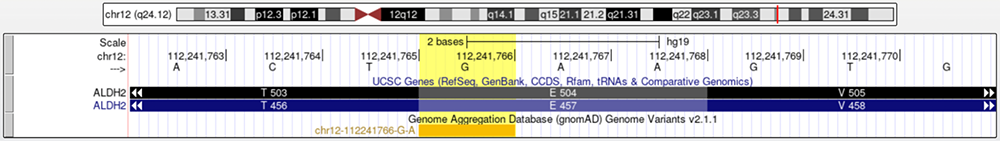
Figure 13. The gnomAD track is enabled and has a marker in rs671'ls position.
 https://genome.ucsc.edu/s/education/hg19_ALDH2gnomAD
https://genome.ucsc.edu/s/education/hg19_ALDH2gnomAD
Clicking into the orange box will take you to gnomAD's details page for this variant (Figure 14). Here, we can see a variety of different populations and their allele frequencies for the rs671 position. Boxed in red are the data regarding "East Asian" populations. As one can see under the "Allele Frequency" column, East Asians have the highest allele frequency (26.7%) compared to the other populations, which are quite low. "Latino/Admixed American" populations have the second highest frequency (0.1%), after "Other"; This may be because the first peoples to populate the Americas crossed over the Bering Strait from Asia, springing some genetic similarities.
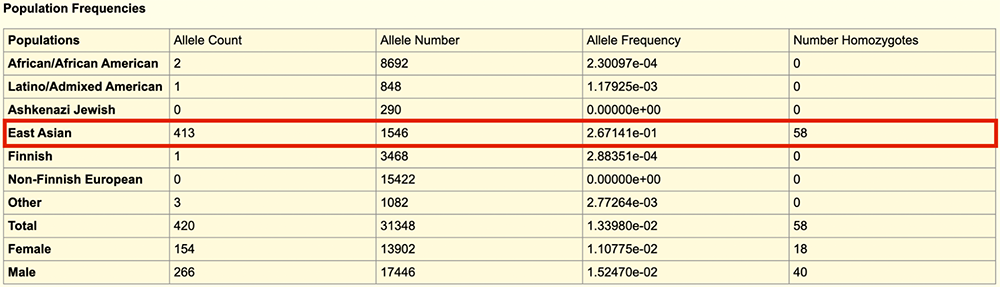
Figure 14. A section of the gnomAD details page for rs671, providing population frequencies.
 link.
link.
Next, let's enable the HGDP track to see what information it can provide. The Human Genome Diversity Project shows allele frequencies for isolated populations in many parts of the world — useful for tracking human migrations. While there is no HGDP marker on this specific nucleotide coordinate, there are rsIDs nearby. These variants are not causative of alcohol flush syndrome like rs671, but sometimes nearby variants will cosegregate; A variant non-causative of the phenotype in question may be passed down in conjunction with the nearby causative variant, therefore still holding relevant information concerning the haplogroup or phenotypic population frequencies, while not actively contributing to the phenotype. Only one of the six variants shown on the HGDP track in Figure 15 (rs4767944, highlighted in dark blue) is confirmed to cosegregate with rs671. Figure 15 shows where rs4767944 lies on ALDH2 in relation to the position of rs671 (highlighted in orange), and Figure 16 shows a close up of rs4767944's position.
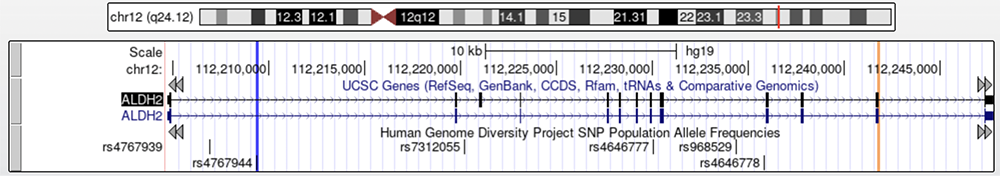
Figure 15. ALDH2 with the HGDP track enabled. The left blue highlight is rs4767944, the HGDP allele in question, while the right orange highlight is rs671, the causative variant of alcohol flush syndrome.
 https://genome.ucsc.edu/s/education/hg19_ALDH2hgdp
https://genome.ucsc.edu/s/education/hg19_ALDH2hgdp
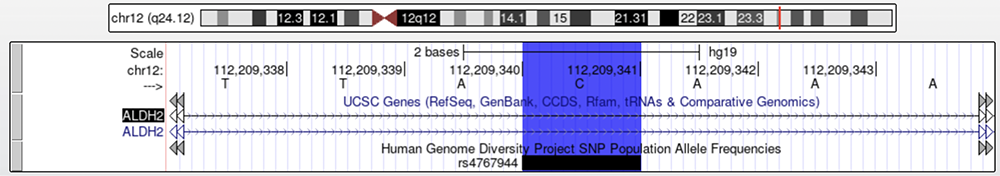
Figure 16. A closeup view of rs4767944, which is located in an intron. The nucleotide in this position has been observed as a "C" or "T."
Clicking into the HGDP marker for rs4767944 will take you to its details page, which includes the image in Figure 17. This image provides a visual representation of the distribution of allele frequencies for this variant. As one can see, the derived allele (T) is found primarily in populations in East Asia and some populations in Northern Asia. It is also observed in some populations in South America (upper right corner). The rest of the world'ls populations exhibit high frequencies of the ancestral allele. Using this data, one can infer that rs4767944 most likely cosegregates with rs671, the causative allele of alcohol flush syndrome.
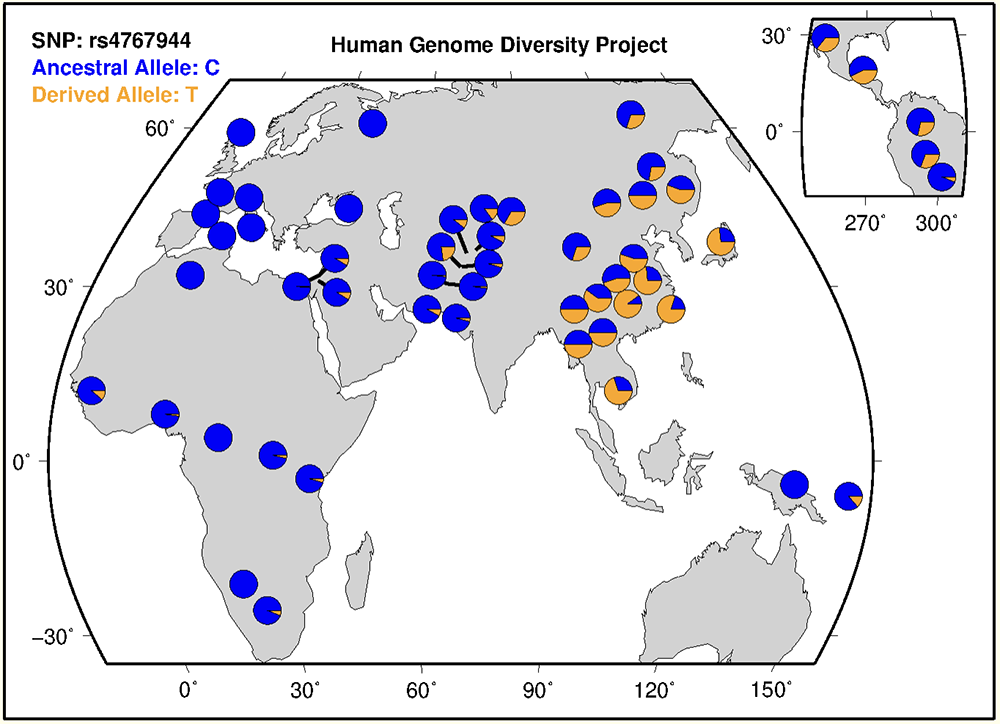
Figure 17. HGDP's allelic population frequencies for rs4767944.
 link.
link.
The allelic population frequencies of rs4767944 shown in Figure 17 are incredibly similar to the gnomAD allelic population frequencies of rs671 (Figure 14), with East Asians having the highest frequency of the derived allele. This can be further corroborated by the gnomAD data (Figure 14) which shows that the "Latino/Admixed American" population has the third highest allele frequency (after "Other"), similar to how the visual data from the HGDP database shows substantial frequencies of the derived allele in South America (Figure 17).
REFERENCES
Lee et al., 2014. Genetic and Sociocultural Factors of Alcoholism Among East Asians
Wikipedia. Alcohol Flush Reaction
Wall and Ehlers., 1995. Genetic Influences Affecting Alcohol Use Among Asians
Written by Zoë Shmidt, UCSC. Major: BA, Biological Anthropology