- Home
- Genomes
- Genome Browser
- Tools
- Mirrors
- Downloads
- My Data
- Projects
- Help
- About Us
LACTASE PERSISTENCE
 https://genome.ucsc.edu/s/education/hg19_LCT
https://genome.ucsc.edu/s/education/hg19_LCT
It is essential that all young mammals are able to break down the main carbohydrate found in their mother's milk, called lactose, into digestible sugars. The enzyme responsible for converting lactose into glucose is called lactase. Lactase is produced by transcribing and translating the LCT gene. Almost all mammalian babies have lactase present in their bodies, allowing them to digest their mothers' milk. As most mammals age and no longer need to rely on milk for nourishment, the LCT gene gradually ceases to be expressed. However, some humans continue to produce lactase throughout their adult lives; this is called lactase persistence, or LP. Populations whose ancestors have historically relied on animal products as their nutritional strategy, such as pastoral, herder, and other agriculturist cultures, tend to have higher frequencies of LP. LP developed within communities independently, meaning each initial occurrence of LP within a population is a result of independent allele changes; European lactase persistence is associated with different allele changes than cases of LP seen in Middle Eastern or African individuals. The genome browser can be used to identify some of the nucleotide changes responsible for prolonging LP into adulthood.
Pictured below is the MCM6 gene found in the human genome (Figure 1); this gene is upstream from the LCT gene and contains nucleotide changes that are associated with lactase persistence. We will be focusing on two SNVs found in MCM6; one is associated with LP in European individuals while the other is thought to be one of the many SNVs linked to African and Middle Eastern LP. Both are found in introns within MCM6.
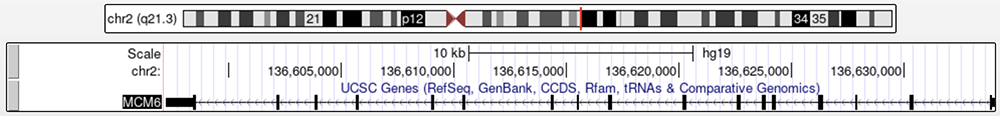
Figure 1a. The MCM6 gene, which is upstream from the LCT gene.
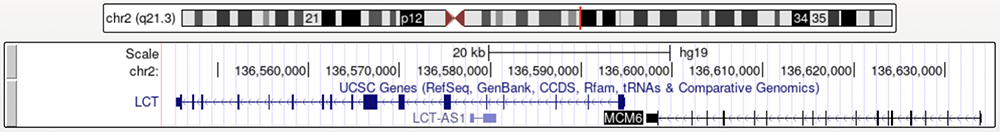
Figure 1b. The MCM6 gene in relation to the LCT gene.
 https://genome.ucsc.edu/s/education/hg19_LCTandMCM6
https://genome.ucsc.edu/s/education/hg19_LCTandMCM6
The Enattah et al. 2002 paper mentions the two variants we will be exploring. The "two variants from Enattah etal 2002" custom track (Figure 2) marks where the SNVs lie on MCM6. The turquoise highlighted vertical line crosses through the -13910 C/T allele change in intron 13 on MCM6 mentioned in the paper. The yellow highlighted vertical line passing through the -22018 mark on the custom track is the G/A allele change on intron 9 mentioned in the same paper. The allele coordinates were named relative to the ATG of the LCT gene.
Both of these highlighted lines also cross through rsIDs (reference SNPs) on the GWAS and OMIM tracks. The GWAS track, or Catalog of Published Genome-Wide Association Studies, is a data track that marks sections of the genome that cosegregate with a phenotype using a convenient, usually non-causative, reference allele. The GWAS track does not necessarily tag causative alleles with rsIDs; rather, it marks convenient alleles that cosegregate with haploblocks associated with a certain phenotype. The Online Mendelian Inheritance in Man, or OMIM track, is a database that catalogs genes that have been published as having the potential to result in genetic disorders or traits.
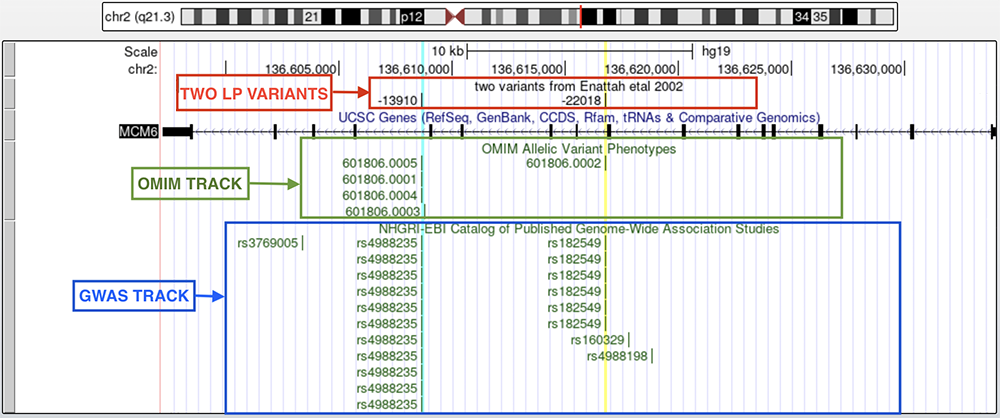
Figure 2a. MCM6 with two highlighted variants. A custom track labeled "two variants from Enattah etal 2002" show the coordinates, relative to the translation start of the LCT gene, of the variants in question. The "OMIM" and "GWAS" tracks are found below the "UCSC Genes" track, in respective order.
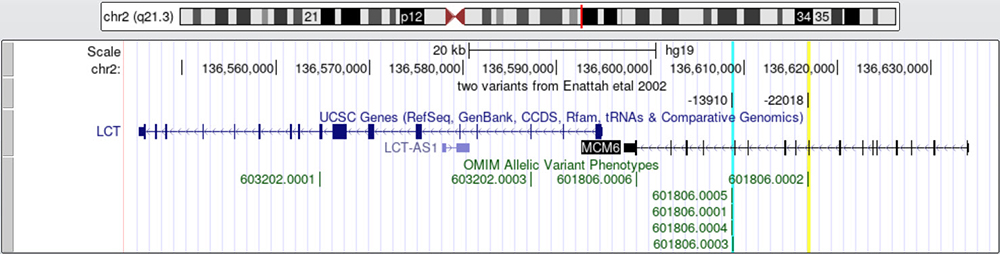
Figure 2b. MCM6 with two highlighted variants. Shows the
relationship of the variants in MCM6 which regulate the nearby LCT gene.
 https://genome.ucsc.edu/s/education/hg19_LCT_MCM6
https://genome.ucsc.edu/s/education/hg19_LCT_MCM6
VARIANT -13910
Figure 3 shows the -13910 change on a base pair level. This is the C>T allele change associated with European LP mentioned in Enattah et al., but the base highlighted on the reference genome is G; this is because we are looking at the complementary strand. The Enattah et al. paper is providing the coordinate from the point of view of transcription (5' > 3'), but the LCT and MCM6 genes are on the bottom strand. Clicking on the boxed arrow to the left of the nucleotide sequence switches the strand and shows the C reference allele. Figure 4 shows what the browser looks like when displaying the bottom strand.
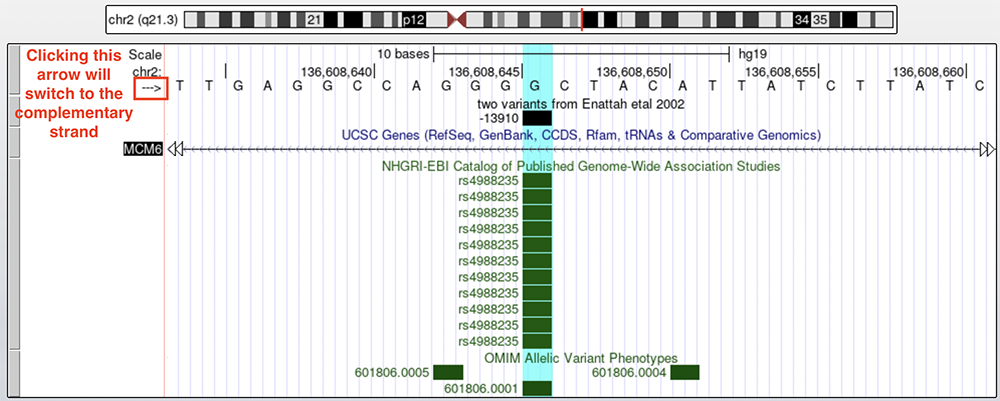
Figure 3. The -13910 allele on the top strand of the genome assembly. Clicking on the boxed arrow will switch to the complementary strand.
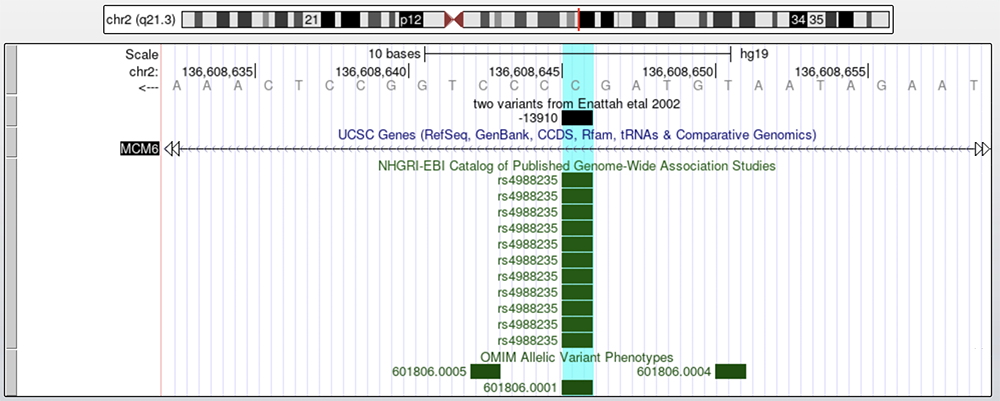
Figure 4. The -13910 allele from the bottom strand perspective of the gene. The nucleotides turn from black to light grey to signify that the complementary strand is being shown.
Clicking into the OMIM item labeled 601806.0001, confirms that this SNV is linked to lactase persistence (Figure 5). The dbSNP/ClinVar line corroborates that this is the same rsID, and therefore variant, seen on the GWAS track.

Figure 5. This is the top of the details page brought up when the OMIM allele variant 601806.0001 is selected from the genome browser page.
Clicking on the linked text "601806.0001" will bring you to the OMIM database, specifically a page detailing this SNV's association with lactase persistence (Figure 6). This page cites the Enattah et al. publication previously mentioned as the research that confirmed -13910's involvement in LP. -13910"s causation of LP is confirmed through this OMIM page; this variant "affects a binding site of transcription factor AP-2" and, therefore, plays a pivotal role in an adult individual's ability to digest lactose.
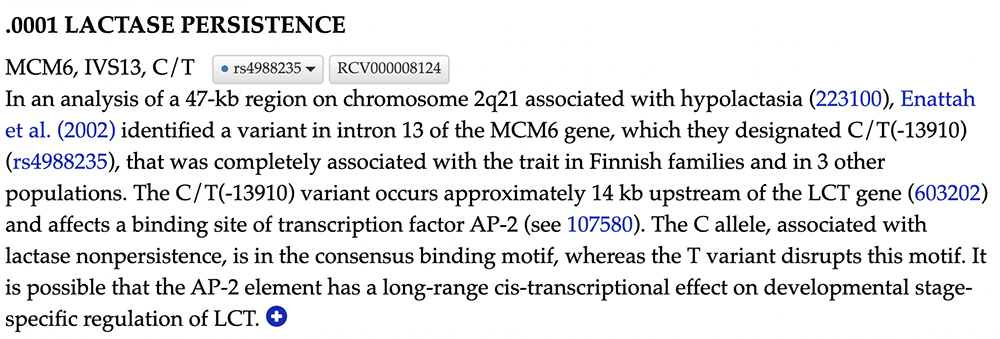
Figure 6. Selecting the linked text "601806.001" will bring the user to OMIM'l;s website and the data page on the variant in question.
Going back to the genome browser and clicking into the GWAS rsID, labeled rs4988235 (Figure 7), will bring up publications that provide more information on the phenotypic differences associated with the allele change and the cosegregating sequence. The Hughes et al. publication references the -13910 variants association with LP in regards to their research. They mention -13910 as, "a variant associated with lactose intolerance in ClinVar and OMIM (#223100), which functions as an enhancer of the LCT gene promoter." While a GWAS rsID does not necessarily point to causation, -13910 is confirmed to be a LP causative variant as reported by Enattah et al. and annotated by OMIM.

Figure 7. The GWAS details page for rs4988235, also known as -13910. This page can be accessed by selecting the rsID from the genome browser.
VARIANT: -22018
Figure 8 shows the -22018 G to A SNV on a basepair level. Once again, this screenshot is of the complementary strand, hence why a "C" nucleotide is highlighted. As before, clicking the boxed gray arrow will switch the reference genome to the opposite strand and the complement bases. The -22018 SNV is thought to be one of the many responsible for lactase persistence in African and Middle Eastern individuals.
This variant also intersects with both an OMIM and GWAS rsID. However, this variant is not confirmed to be causative; rather, it is included within a cosegregating haploblock associated with the phenotype, lactase persistence. We will explore causation further when discussing -22018's GWAS rsID.
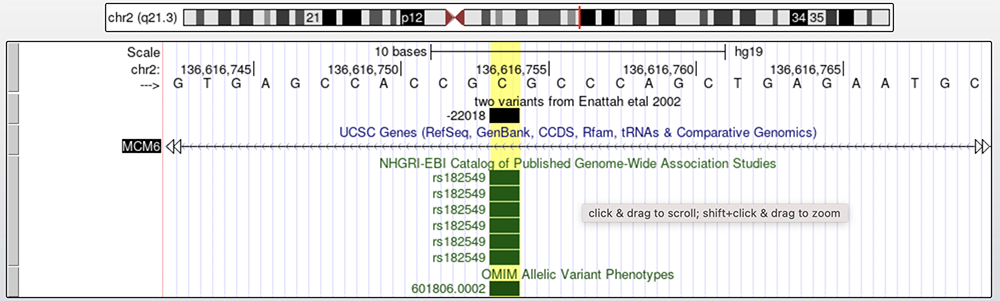
Figure 8. The -22018 allele is found on the "two variants from Enattah etal 2002" track. Below the "UCSC Genes" track, which shows a section of intron 9 on MCM6, is the "GWAS" track and the "OMIM" track, respectively.
Clicking into the OMIM rsID (Figure 9), 601806.0002, confirms that this SNV is also linked to lactase persistence, just as the last SNV. The dbSNP/ClinVar line corroborates that this is the same rsID, and therefore variant, seen on the GWAS track.

Figure 9. This is the top of the details page brought up when the OMIM allele variant 601806.0002 is selected from the genome browser page
Exploring the OMIM rsID (Figure 10) provides similar results to the first variant. The OMIM data page on the rsID also uses the Enatah et al. publication to confirm this SNV's role in lactase persistence. However, nothing in this OMIM excerpt points to confirmed causation in the same way as the -13910 variant. Instead, this variant is a part of a "lactase persistence-associated haplotype", meaning the variant cosegregates with a haploblock that is associated with LP rather than being a causative allele itself.
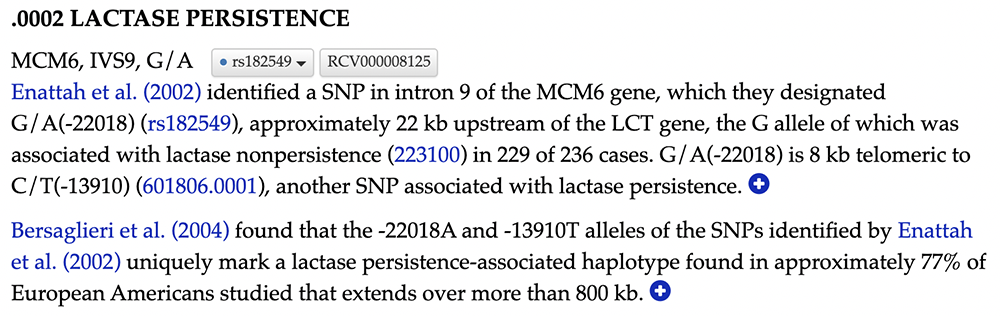
Figure 10. Selecting the linked text "601806.002" will bring the user to OMIM's website and the data page on the variant in question.
Navigating back to the genome browser and clicking into the GWAS rsID for the variant labeled rs182549 brings one to another page of information and publications (Figure 11, 12). These publications referencing "glucose metabolism" and "human gut microbiome composition" as traits linked to the allele both mention the -13910 and -22018 variants" role in LP. The links to the publications themselves provide more information on the phenotypic differences associated with the variants. However, none of the publications found within the GWAS rsID nor the OMIM database point to -22018 being confirmed as a causative allele; rather, the SNV is included within a cosegregating haploblock and acts as a convenient marker for the block as a whole.

Figure 11. The GWAS details page for rs182549, also known as -22018, showing association of the variant with glucose metabolism. This page can be accessed by selecting the highlighted rsID on the GWAS track from the genome browser.

Figure 12. The GWAS page for rs182549, also known as -22018, continued. This report, from a different study, associates the SNV with gut microbiome composition.
REFERENCES
Enattah, et al., 2002. Identification of a variant associated with adult hypolactasia
Hughes, et al., 2020. Genome-wide associations of human gut microbiome variation
Evolution of lactase persistence
Genome-wide association study of genetic loci linked to glucose metabolism
Identify host factors influencing human gut microbiome composition
Written by Zoë Shmidt, UCSC. Major: BA, Biological Anthropology