- Home
- Genomes
- Genome Browser
- Tools
- Mirrors
- Downloads
- My Data
- Projects
- Help
- About Us
EXPLORING CAG AND CAA POLYGLUTAMINE REPEATS
CAG repeats are repeated sequences encoding anywhere from 6-37 glutamine amino acids. In this session, we will be using the information learned from the discussion of CAG Repeats in Huntington's Disease and applying it to search the entire human genome for CAG repeats. In doing so, we will observe several genes and analyze each for similarities.
The Genome Browser tool we will be using to aid our search for CAG repeats across the genome is the "Short Match" data track. "Short Match" searches a section of the genome displayed on the browser graphic for sequences matching the sequence entered on the configuration page. In Figure 1 we can see where the "Short Match" tool is located on the browser. In Figure 2 the "Short Match" track configuration page can be seen with a search sequence typed into the text box (Figure 2).
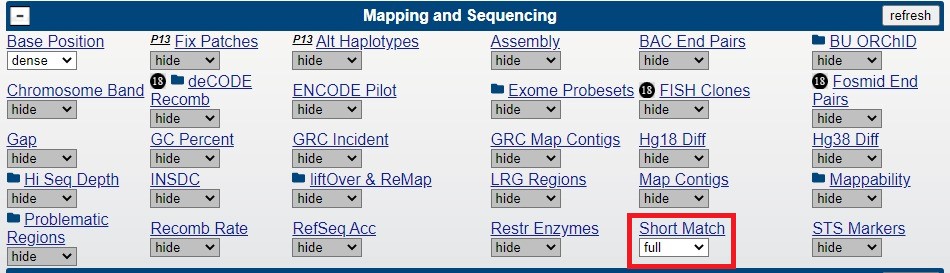
Figure 1. "Short Match" feature location on the main session page (under "Mapping and Sequencing" section).
We will search for CAG repeats but include CAA codons, which also encode glutamine, for this demonstration using the "Short Match" tool. We can do this by literally typing out CAG several times followed by CAA or just typing CAR out several times, where R is an unspecified purine nucleotide that can be either A or G (Figure 2). In this instance, we will type CAR out eight times to find runs of polyglutamine of eight amino acids or more.
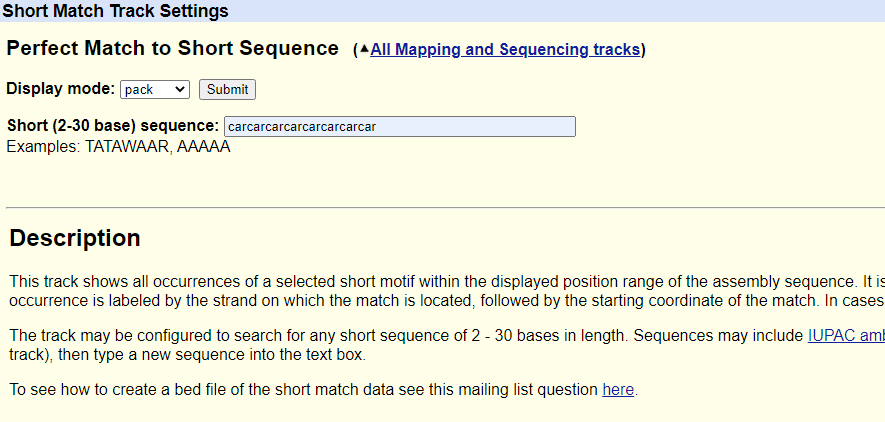
Figure 2. Typed-out search query sequence in the "Short Match" tool.
 Short Match configuration.
Short Match configuration.
By clicking on the submit button, "Short Match" will analyze all the nucleotide sequences present in the current browser window and highlight any matches or "hits". To demonstrate this, starting on the Browser, with the "Short Match" track set to find CAR repeats, we can look at large regions of a chromosome and locate other genes with this motif. Scanning chromosome X, we find a gene MED12 with a polyglutamine region (Figure 4). On the gene details page, we can see that "the MED12 protein is essential for activating CDK8 kinase," and Opitz-Kaveggia syndrome, which are associated with neurological phenotypes.
With the "Short Match" track, we see all the CAG repeats near the 3' end of the MED12 gene (Figure 3). Zooming in on this exon will bring us to Figure 4, showing that the hits of polyglutamine are in exons 42 and 43.
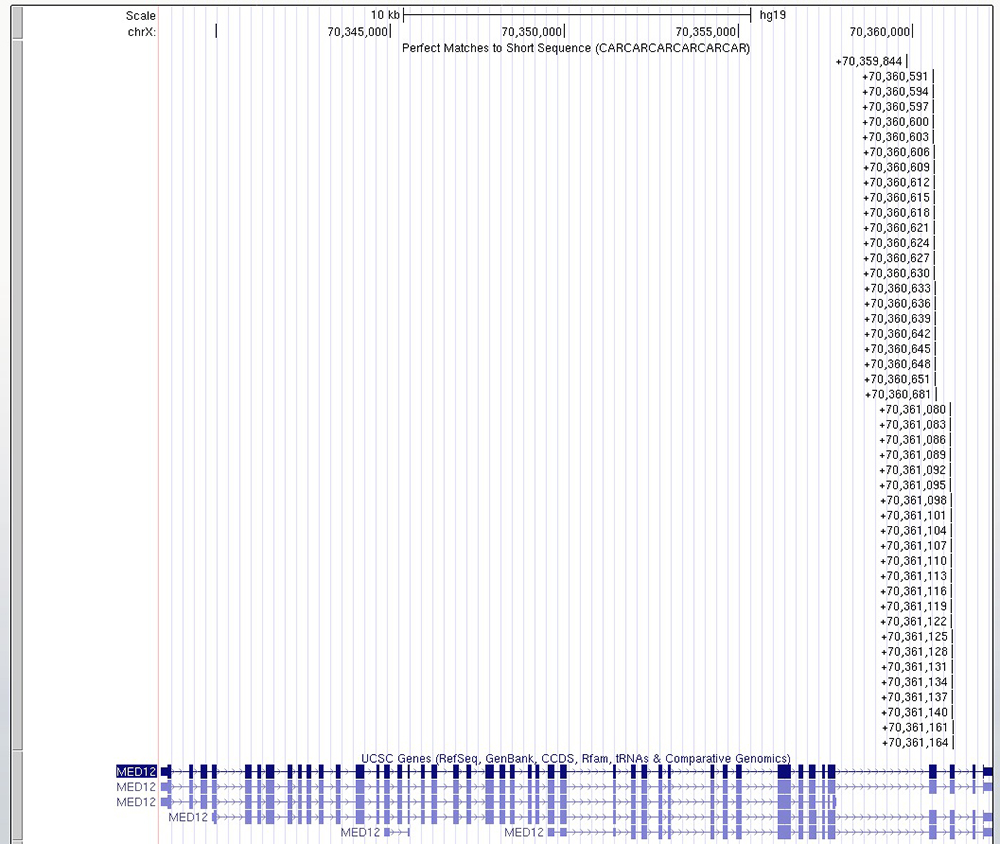
Figure 3. Using the "Short Match" tool on the Browser and typing in our search query sequence (seen in Figure 2), we find multiple hits for CAG/CAA repeats near the 3' end of the MED12 gene.
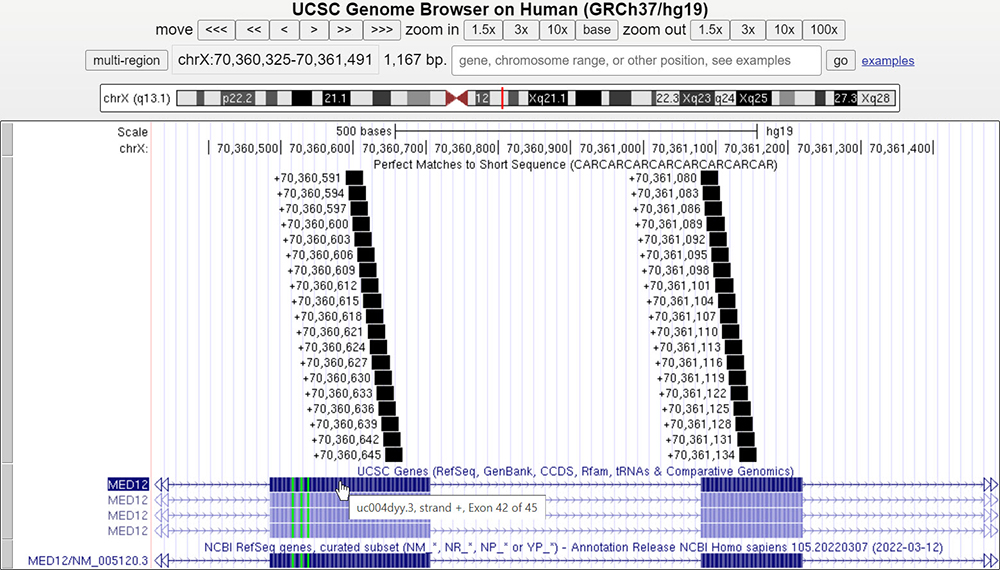
Figure 4. A closer examination of exons 42 and 43 shows our "Short Match"
CAR hits localized at the two exons.
 https://genome.ucsc.edu/s/education/anom_fig6
https://genome.ucsc.edu/s/education/anom_fig6
If we zoom in even closer individually on exons 42 and 43, we can directly count the number of repeats. In Figure 5, we can observe that exon 42 has 26 glutamines in a row, and in Figure 6, we can observe that exon 43 also has 26 glutamines in a row. Something worth noting is that after both repeat sequences, a smaller repeat sequence follows. This can be seen in Figure 5 marked in orange.
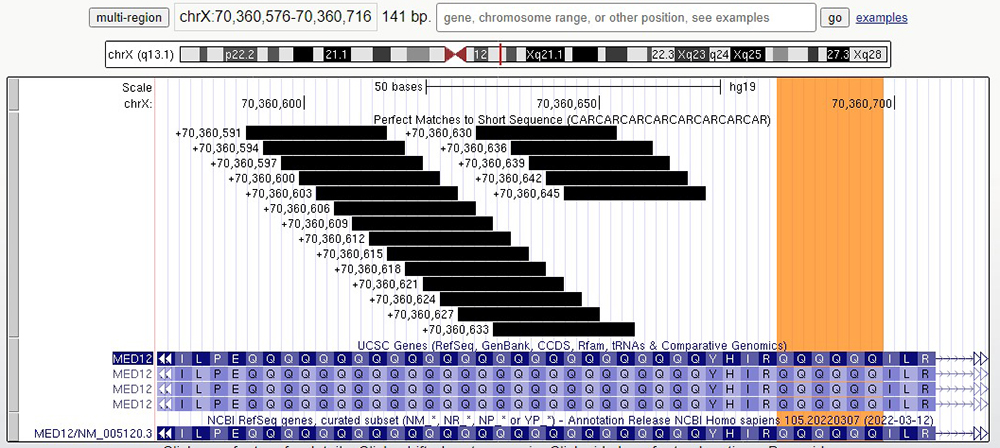
Figure 5. Zoom-in on exon 42, displaying 26 glutamine residue followed by a short repeat sequence of six glutamines highlighted in orange following the main repeat sequence.
 https://genome.ucsc.edu/s/education/anom_fig7
https://genome.ucsc.edu/s/education/anom_fig7
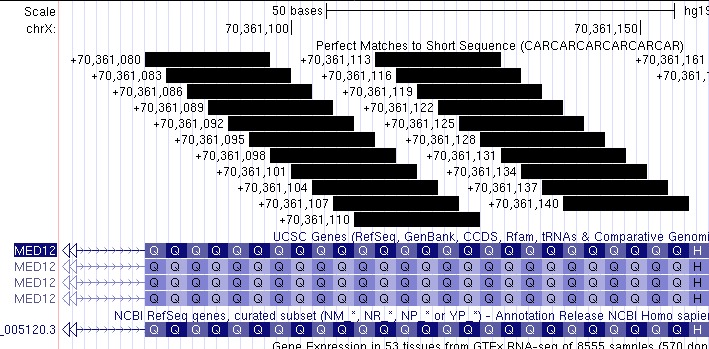
Figure 6. Zoom-in on exon 43 showing 26 glutamine repeats. In comparison to the repeats in Figure 5, these repeats start from the beginning of the exon instead of halfway through.
Comparing the gene MED12 to the HTT gene [link: cag.html], one similarity can be found: both are responsible for medical disorders in cases where there is an expansion of the CAG repeats. For the HTT gene, Huntington's disease, and for MED12, FG syndrome, an X-linked genetic disorder that is characterized by poor muscle tone (hypotonia), intellectual disability, constipation and/or anal anomalies, and complete or partial absence of the part of the brain that connects the two hemispheres. Two genes with noticeable sections of CAG repeats have the potential to create brain defects in a patient.
Interestingly, these two genes are not the only genes containing CAG repeat sequences associated with brain defects or body disorders. Another gene, RUNX2, contains a series of 23 glutamine amino acids (Figure 7). RUNX2 plays an important role in skeletal gene expression and mutations in this gene have been associated with the bone development disorder cleidocranial dysplasia (CCD). Like MED12, mutations in RUNX2 result in phenotypes throughout the body, caused by an expansion of CAG repeats.
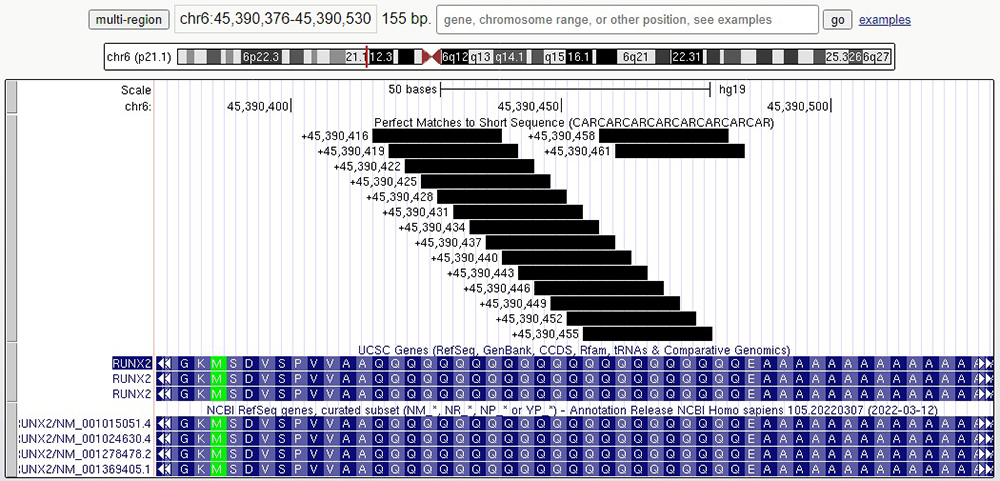
Figure 7. Exon of RUNX2 gene containing CAG/glutamine repeat sequence. In this gene, the repeat stretches for 23 amino acids.
 https://genome.ucsc.edu/s/education/anom_fig9
https://genome.ucsc.edu/s/education/anom_fig9
One last example gene to illustrate this phenomenon is the gene AR (Figure 8), responsible for coding a protein that has three major functional domains: the N-terminal domain, DNA-binding domain, and androgen-binding domain. In AR, there are 23 CAG repeats followed by a small section of six repeats a few amino acids further downstream. As indicated on the gene's details page, "Expansion of the polyglutamine tract from the normal 9-34 repeats to the pathogenic 38-62 repeats causes spinal bulbar muscular atrophy (SBMA, also known as Kennedy's disease)." Kennedy's disease is a disorder of specialized nerve cells that control muscle movement (motor neurons). These nerve cells originate in the spinal cord and the part of the brain that is connected to the spinal cord (the brainstem). Once again, a "pathogenic" number of expanded CAG repeats is responsible for a disease correlating to the brain and spine, similar to the genes discussed above.
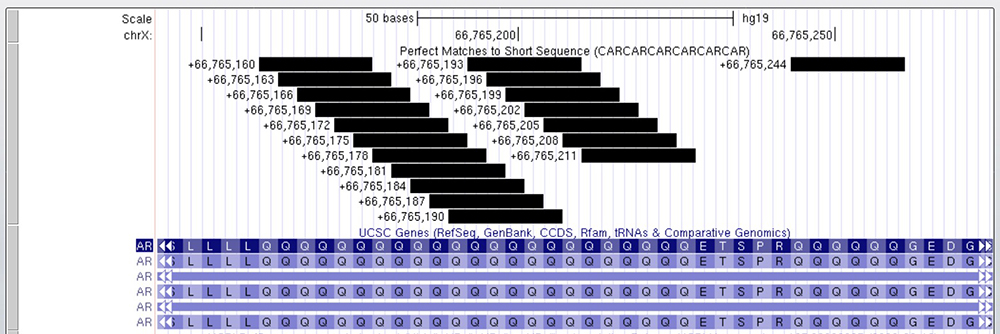
Figure 8. Exon within the AR gene with a series of 23 CAG repeats. Following this section of repeats is a smaller section of repeats with six glutamine repeats, a pattern which was seen in the MED12 as well. The Short Match in this case identifies runs of 6 Glns.
These genes are not the only genes with this motif. Other genes, including ATN1, BEAN1, AFF2 and C9ORF72 all contain long CAG repeats and all contribute to similar medical disorders and diseases.
A number of mechanisms have been proposed to explain the pathogenic effect of CAG-repeat expansion Is the mechanism at the DNA or RNA level (and hence CAG and CAA may have different effects), or is it at the protein level (and therefore CAG and CAA would be equivalent)? Why are so many of the genes with CAG repeats expressed in the brain or nervous system?
Written by Mateo Etcheveste, UCSC. Major: BS, Biomolecular Engineering